|
Ordering information |
|||
|
Name |
Cat. No. |
Vol. |
Scheme |
|
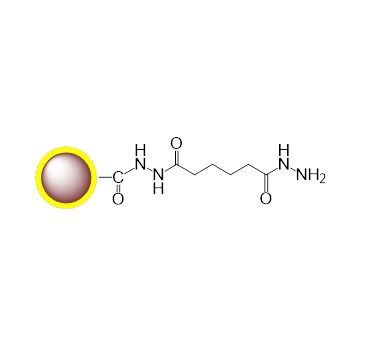
G-Hydrazide |
PMG042-2 PMG042-5 |
2 ml 5 ml |
|
1. Overview
PuriMag™ G-Hydrazide, Hydrazide-Activated Magnetic Nanoparticles are uniform, polymer-coated superparamagnetic nanoparticles. Their hydrophilic surface ensures excellent dispersibility, low non-specific adsorption, and easy handling in diverse buffers. The beads feature a high-density surface coating of hydrazide functional groups, enabling site-specific covalent immobilization of aldehyde/ketone-containing ligands (e.g., antibodies, lectins, glycoproteins, carbohydrates).
Key Applications:
Glycoprotein immobilization via periodate-oxidized carbohydrate aldehydes
Ideal for polyclonal antibody coupling through Fc-region carbohydrates, preserving antigen-binding orientation
Compatible with carbohydrate-rich monoclonal antibodies
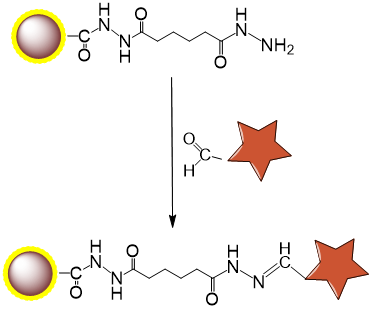
Immobilization Chemistry:
Periodate oxidation of cis-diol groups in glycans generates aldehydes.
Spontaneous reaction between aldehydes and bead hydrazide groups forms stable hydrazone bonds.
Features & Advantages:
Efficient covalent coupling
Stable hydrazone bonds with minimal ligand leakage
Generates reusable immunoaffinity matrices
Low non-specific binding
Capacity: 1–20 mg protein or 0.1–2 mg peptide per gram beads
Long hydrophilic spacers minimize steric hindrance
Operational Flexibility:
Adaptable coupling time/temperature
Compatible with affinity chromatography workflows
Schematic Diagram of Bead Coupling Mechanism:
2. product description
Product Specifications
Description
Polymer coated Fe3O4 nanoparticles
Particle Size
200 nm
Number of Beads
~1.7×1010 beads/mg
Matrix
Proprietary polymer
Functional group
Hydrazide groups
Group density
~300 µmole / g of Beads
Magnetization
60~70 EMU/g
Formulation
10mg/mL in in 1M NaCl and 0.02% NaN3
Stability
pH 3.5~10, 4~50 ℃, most organic solvents
Storage
1 year at 2~8 ℃. Do not freeze.
3. Instructions for Use
Note: This protocol exemplifies coupling proteins/peptides to PuriMag™ G-Hydrazide beads. Titration-based bead optimization for individual applications is strongly recommended. Scale proportionally. Avoid Tris or buffers containing primary amines/sugars, as they compete with coupling.
A. Required Materials
1.Magnetic Racks:
For 12×1.5–2 mL tubes (Mrack02)
For 2×50 mL or 2×15 mL tubes (Mrack03)
2.Coupling Buffer: 0.1 M Sodium Phosphate, pH 7.0 or 0.1 M Sodium Acetate, pH 5.6
3.Oxidizing Agent: Sodium meta-Periodate (NaIO₄)
4.Desalting Column: Sephadex G-25
5.Wash Buffer: 1 M NaCl
B. Protein Coupling
B-1. Glycoprotein Oxidation
Light-sensitive reaction – perform in dark.
1.Dissolve/dilute 0.5–10 mg glycoprotein in 1 mL coupling buffer.
(If pre-suspended in other buffers, exchange via dialysis/desalting.)
2.Add protein solution to amber vial containing 2 mg NaIO₄ (10 mM final). Gently agitate to dissolve oxidizer.
3.Incubate with gentle agitation at RT in dark for 30 min.
4.Terminate reaction. Remove unreacted NaIO₄ via Sephadex G-25 desalting/buffer exchange:
Equilibrate column with 5 mL coupling buffer.
Load oxidized sample onto column bed.
Wash with 0.5 mL coupling buffer into bed.
Elute with 2 mL coupling buffer; collect eluate (typically 1.5–2 mL).
B-2. Coupling to Hydrazide-Terminated Beads
*Note: Store beads as 30 mg/mL suspension in 1 mM EDTA at 4°C. Vortex vigorously before use.*
1.Transfer 0.5 mL fully resuspended beads (30 mg/mL) to microcentrifuge tube.
2.Place tube in magnetic separator for 1–3 min. Discard supernatant while tube remains in separator.
3.Remove tube. Resuspend beads in 1.5 mL coupling buffer.
4.Repeat Steps 2–3 three times (total 4 washes).
5.Resuspend beads in 750 μL coupling buffer.
6.Add 250 μL oxidized protein solution. Incubate at RT for ≥6 h.
*Note: Optimize protein concentration empirically. For 10 mg beads, start with 100–250 μg/mL.*
7.Wash beads 3× with 1 mL wash buffer.
8Wash beads 3× with 1 mL desired storage buffer.
9.Resuspend in PBS with 0.1% sodium azide (w/v). Store at 4°C (avoid freezing).
C. General Affinity Purification Protocol
Note: Protein purification requires application-specific optimization due to structural diversity.
1.Transfer optimized bead amount to centrifuge tube. Perform magnetic separation for 1–3 min. Discard supernatant.
Note: Titrate beads against target protein abundance. Typical binding: 1–20 μg target protein per mg beads. Excess beads increase background; insufficient beads reduce yield.
2.Wash beads 3× with 5 bead volumes PBS:
Remove tube from magnet.
Resuspend in PBS for 30 sec.
Separate magnetically (1–3 min). Discard supernatant.
3.Add washed beads to crude sample containing target protein. Incubate 1–2 h at RT or optimized temperature (extend for lower temperatures).
4.Wash with PBS or 1M NaCl until eluate OD₂₈₀ < 0.05.
5.Elute target using:
Low pH (2–4)
High pH (10–12)
High-salt buffer
Heat denaturation
Affinity elution
Boiling in SDS-PAGE loading buffer
(For research use only!)