江南大学采用我司链霉亲和素磁珠进行MB-SELEX通过12轮筛选筛选出阿奇霉素特异性适配体
江南大学采用我司链霉亲和素磁珠进行MB-SELEX通过12轮筛选筛选出阿奇霉素特异性适配体
In vitro selection and engineering azithromycin-specific aptamers and construction of a ratiometric fluorescent aptasensor for sensitive detection of azithromycin
Tianyu Huang, Xin Chen, Jinri Chen, Yuting Zhang, Xiaoli Wang, Zhimeng Wu, Nandi ZhouThe Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, China
Received 12 December 2023, Revised 7 April 2024, Accepted 7 April 2024, Available online 9 April 2024, Version of Record 9 April 2024.
Sensors and Actuators B: Chemical
Volume 411, 15 July 2024, 135789
https://doi.org/10.1016/j.snb.2024.135789
Highlights
• DNA library-immobilized MB-SELEX was used to screen azithromycin-specific aptamers.
• Truncating non-binding segments of aptamers was based on structural prediction and rational design.
• Specific single-base mutations were based on molecular docking analysis.
• The aptamer was used in a ratiometric fluorescent aptasensor for azithromycin.
• The aptasensor has been successfully utilized for detection of azithromycin in actual samples.
Abstract
Azithromycin can effectively inhibit bacterial protein synthesis and reduce biofilm formation and quorum sensing, and thus has been preferentially used in the treatment of various infections. However, azithromycin is also among the most highly concentrated antibiotics present in wastewater due to the inappropriate use and treatment. In order to sensitively and rapidly monitor the azithromycin residues, aptamers specific to azithromycin were selected through 12 rounds of screening by using magnetic beads-SELEX (MB-SELEX). The screened aptamers were assessed for their affinity and specificity using fluorescent assay. Apt3 emerged as the optimal candidate aptamer with the dissociation constant (Kd) of 235.07 nM. After secondary structure analysis and molecular docking, Apt3 was systematically truncated and subjected to single-base mutations. The optimal aptamer Apt3–27 T was identified, exhibiting the Kd value of 215.84 nM and the improved specificity compared to the original aptamer. Then Apt3–27 T was utilized to construct a ratiometric fluorescent aptasensor, which exhibited the linear dynamic range of 25–400 nM and low detection limit of 9.78 nM. More importantly, the ratiometric fluorescence mechanism further improved the specificity of the biosensor, which finally can discriminate azithromycin from structurally similar erythromycin. Evaluation experiments using actual samples spiked with azithromycin showed the recovery ranging from 97.1% to 106% and the relative standard deviation ranging from 4.29% to 8.61%. The screened aptamer and constructed aptasensor offer the advantages of high affinity, high effectiveness and ease of use in actual sample detection, thereby displaying promising applications in the monitoring of azithromycin residues.
阿奇霉素能有效抑制细菌蛋白质合成,减少生物膜形成和群体感应,因此被优先用于各种感染的治疗。然而,由于使用和处理不当,阿奇霉素也是废水中浓度最高的抗生素之一。为了灵敏、快速地监测阿奇霉素残留,采用磁珠-SELEX(MB-SELEX)通过12轮筛选筛选出阿奇霉素特异性适配体。使用荧光测定法评估筛选的适配体的亲和力和特异性。Apt3 成为最佳候选适配体,解离常数 (Kd)的235.07 nM。经过二级结构分析和分子对接,Apt3被系统地截断并发生单碱基突变。确定了最佳适配体 Apt3–27 T,表现出 Kd值为 215.84 nM,与原始适配体相比,特异性有所提高。然后利用Apt3–27 T构建了比例式荧光适配体传感器,其线性动态范围为25–400 nM,检出限低至9.78 nM。更重要的是,比例荧光机制进一步提高了生物传感器的特异性,最终可以区分阿奇霉素和结构相似的红霉素。使用阿奇霉素加标的实际样品的评估实验显示,回收率在97.1%至106%之间,相对标准偏差在4.29%至8.61%之间。筛选的适配体和构建的适配体传感器在实际样品检测中具有亲和力高、有效性和易用性等优点,在阿奇霉素残留监测中具有广阔的应用前景。
Graphical Abstract
In vitro selection and engineering azithromycin-specific aptamers and construction of a ratiometric fluorescent aptasensor for sensitive detection of azithromycin
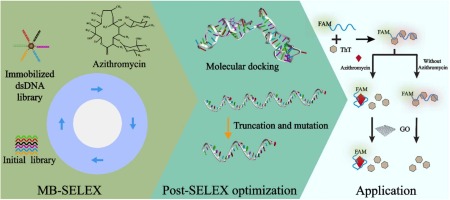
2. Experimental section
2.1. Materials and reagents
Azithromycin, erythromycin, roxithromycin, clarithromycin, streptomycin, penicillin and chloramphenicol were purchased from Macklin Biochemical Co., Ltd (Shanghai, China). DNA oligonucleotides were synthesized by Sangon Biotech Co., Ltd (Shanghai, China). The sequences are listed in Table S1. PCR and electrophoresis reagents were obtained from TaKaRa Biotechnology Co., Ltd (Dalian, China). Streptavidin-modified magnetic beads were purchased from PuriMag Biotechnology Co., Ltd (Xiamen, China). GO (20 mg/mL) was obtained from Aladdin Biochemical Technology Co., Ltd (Shanghai, China). SYBR Green I (SGI, 10000×) was from Solarbio Science & Technology Co., Ltd. (Beijing, China). All other chemicals were purchased from Sinopharm Chemical Reagents Co., Ltd. (Shanghai, China). Ultrapure water (18.2 MΩ cm) obtained from a Millipore water purification system (Gradient A10, Merck Millipore, Burlington, USA) was used in all experiments. Lake water was collected from Taihu Lake, Wuxi, China. Milk was purchased from local market. All antibiotics and sequences were diluted with binding buffer (8 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, pH 7.4).
2.2. Preparation of the immobilized dsDNA library
The initial ssDNA library modified with 6-FAM (1 μM) was amplified using the forward primer modified with 6-FAM fluorophore and the reverse primer modified with biotin (The PCR reaction system is detailed in Table S2). The PCR cycling conditions were as follow: initial denaturation at 95 ℃ for 5 min, followed by 4 cycles of denaturation, annealing and extension at 95 ℃ for 15 s, 55 ℃ for 15 s, and 72 ℃ for 15 s, respectively. Subsequently, 95 μL PCR product was mixed with 105 μL binding buffer. 0.5 mg streptavidin-modified magnetic beads were then added to the mixture and incubated at 25 ℃ for 1 h with continuous shaking. dsDNA was immobilized onto magnetic bead surface through the specific interaction between biotin and streptavidin. Finally, the mixture was washed three times with binding buffer to remove unbound oligonucleotides.
更多链霉亲和素磁珠 链霉亲和素磁珠|SA磁珠|亲和素磁珠|PuriMag G-streptavidin 高biotin结合容量-生物磁珠专家 (purimagbead.com)
- 上一篇:客户采用我司硅磁珠控制药物晶体的多态性筛选在《Journa 2024/5/18
- 下一篇:客户采用PuriMag Si-OH磁珠提取猴痘病毒DNA在《 2024/5/18

